Drawing fragments:
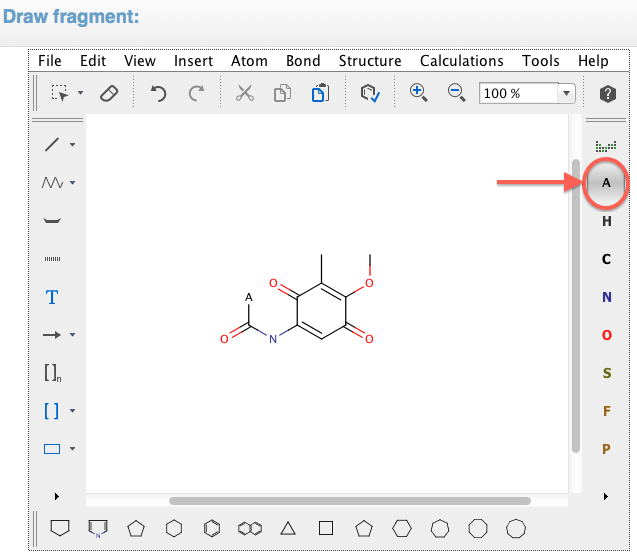
Draw the desired fragment you want to compare into the ChemAxon Marvin Sketcher.
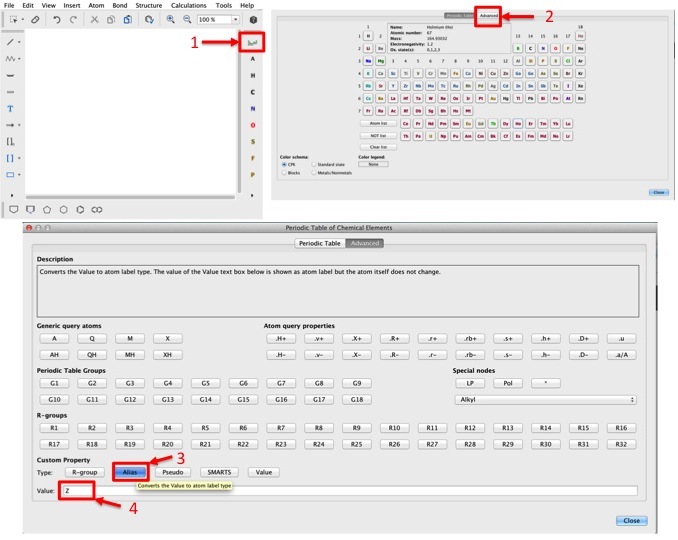
To specify a "linker", use the A atom at the top right corner of the sketcher.
You can also retrieve fragments from a specific PDB ID (i.e. 3ERT) or by entering the protein name (e.g. Thrombin). For the latter the full name is not necessary: "Mitogen-activated protein kinase 14" can
be simplified into "Mitogen-activated" or "kinase 14"
Search Type:
Multiple kind of search type can be performed :
| Substructure : | Finds all molecules containing the given substructure. |
| Superstructure : | Finds all molecules that can be found in the given superstructure. |
| Exact : | Finds molecules that are equal to the query structure (no additional fragments are allowed).
Molecular features (stereochemistry, isotopes, charges, etc.) are by default evaluated the same way
as for substructure queries. See also Perfect search type and Advanced search options. |
| Exact fragment : | Finds molecules where the query exactly matches a fragment of the target.
Other fragments may be present in the target, they are ignored.Useful to perform an Exact
search that ignores salts or solvents beside the main structure in the target. |
| Perfect : | Similar to Exact, but for all molecular features (stereochemistry, charges, isotopes, etc)
equality is needed. It is primarily used for duplicate check. |
In the case where not fragments were found find for your query, an automatic similarity search is performed
Max Hits
Specifies an upper limit for the number of molecules the search engine is allowed to find.
Max Time
Specifies an upper limit for the length of time the search may take.