



2.460 Å
X-ray
2012-03-21
| Name: | Dehydrosqualene synthase |
|---|---|
| ID: | CRTM_STAAU |
| AC: | A9JQL9 |
| Organism: | Staphylococcus aureus |
| Reign: | Bacteria |
| TaxID: | 1280 |
| EC Number: | / |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 100 % |
| B-Factor: | 36.836 |
|---|---|
| Number of residues: | 32 |
| Including | |
| Standard Amino Acids: | 32 |
| Non Standard Amino Acids: | 0 |
| Water Molecules: | 0 |
| Cofactors: | |
| Metals: | |
| Ligandability | Volume (Å3) |
|---|---|
| 0.976 | 610.875 |
| % Hydrophobic | % Polar |
|---|---|
| 60.77 | 39.23 |
| According to VolSite | |
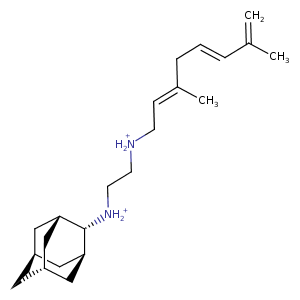
| HET Code: | 3RX |
|---|---|
| Formula: | C22H39N2 |
| Molecular weight: | 331.558 g/mol |
| DrugBank ID: | - |
| Buried Surface Area: | 51.2 % |
| Polar Surface area: | 28.64 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 1 |
| H-Bond Donors: | 2 |
| Rings: | 4 |
| Aromatic rings: | 0 |
| Anionic atoms: | 0 |
| Cationic atoms: | 1 |
| Rule of Five Violation: | 0 |
| Rotatable Bonds: | 9 |
| X | Y | Z |
|---|---|---|
| -5.56242 | -28.381 | 10.1092 |
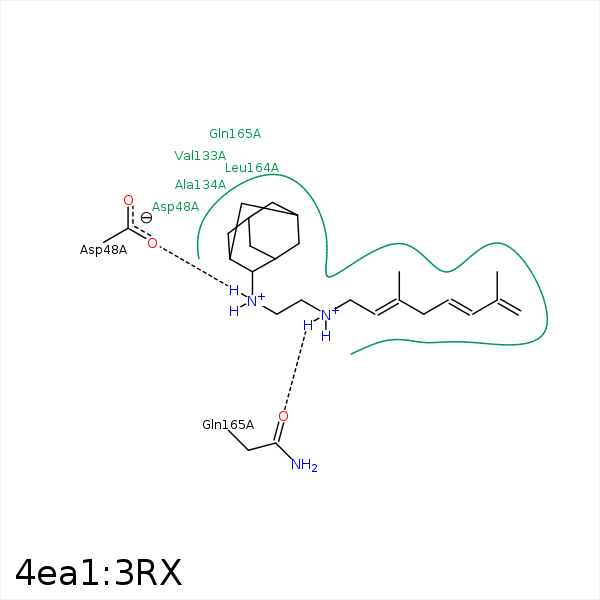
 Image generated by PoseView
Image generated by PoseView



Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| CAC | CE2 | PHE- 22 | 3.84 | 0 | Hydrophobic |
| NAQ | OD1 | ASP- 48 | 2.9 | 135.08 | H-Bond (Ligand Donor) |
| NAQ | OD1 | ASP- 48 | 2.9 | 0 | Ionic (Ligand Cationic) |
| CAO | CB | ASP- 48 | 4.22 | 0 | Hydrophobic |
| CAM | CG2 | VAL- 111 | 3.69 | 0 | Hydrophobic |
| CAL | CB | ASP- 114 | 4.28 | 0 | Hydrophobic |
| CAL | CZ | TYR- 129 | 4.46 | 0 | Hydrophobic |
| CAV | CG2 | VAL- 133 | 3.66 | 0 | Hydrophobic |
| CAJ | CB | ALA- 134 | 3.5 | 0 | Hydrophobic |
| CAJ | CB | VAL- 137 | 4.2 | 0 | Hydrophobic |
| CAA | CD1 | LEU- 141 | 4.42 | 0 | Hydrophobic |
| CAA | CD1 | LEU- 164 | 4.35 | 0 | Hydrophobic |
| CAC | CB | LEU- 164 | 3.99 | 0 | Hydrophobic |
| NAP | OE1 | GLN- 165 | 2.81 | 154.62 | H-Bond (Ligand Donor) |