



1.980 Å
X-ray
2010-05-25
| Min | Mean | Median | Standard Deviation | Max | Count | |
|---|---|---|---|---|---|---|
| pChEMBL: | 7.070 | 7.070 | 7.070 | 0.000 | 7.070 | 2 |
| Name: | Nitric oxide synthase, brain |
|---|---|
| ID: | NOS1_RAT |
| AC: | P29476 |
| Organism: | Rattus norvegicus |
| Reign: | Eukaryota |
| TaxID: | 10116 |
| EC Number: | 1.14.13.39 |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 21 % |
| B | 79 % |
| B-Factor: | 37.191 |
|---|---|
| Number of residues: | 33 |
| Including | |
| Standard Amino Acids: | 27 |
| Non Standard Amino Acids: | 4 |
| Water Molecules: | 2 |
| Cofactors: | |
| Metals: | CL CL ZN |
| Ligandability | Volume (Å3) |
|---|---|
| 0.292 | 2392.875 |
| % Hydrophobic | % Polar |
|---|---|
| 43.72 | 56.28 |
| According to VolSite | |
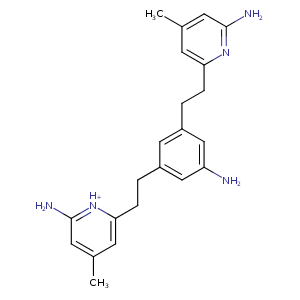
| HET Code: | XFN |
|---|---|
| Formula: | C22H27N5 |
| Molecular weight: | 361.483 g/mol |
| DrugBank ID: | - |
| Buried Surface Area: | 62.14 % |
| Polar Surface area: | 103.83 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 5 |
| H-Bond Donors: | 3 |
| Rings: | 3 |
| Aromatic rings: | 3 |
| Anionic atoms: | 0 |
| Cationic atoms: | 0 |
| Rule of Five Violation: | 0 |
| Rotatable Bonds: | 6 |
| X | Y | Z |
|---|---|---|
| 15.5869 | 2.60733 | 54.4688 |
 Image generated by PoseView
Image generated by PoseView



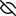
Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| C16 | SD | MET- 336 | 3.56 | 0 | Hydrophobic |
| C38 | CB | PRO- 565 | 3.98 | 0 | Hydrophobic |
| C07 | CG2 | VAL- 567 | 4.13 | 0 | Hydrophobic |
| C36 | CG2 | VAL- 567 | 3.53 | 0 | Hydrophobic |
| C42 | CD1 | PHE- 584 | 3.57 | 0 | Hydrophobic |
| N41 | O | TRP- 587 | 2.86 | 153.58 | H-Bond (Ligand Donor) |
| N40 | OE1 | GLU- 592 | 2.74 | 155.43 | H-Bond (Ligand Donor) |
| N40 | OE2 | GLU- 592 | 3.41 | 127.97 | H-Bond (Ligand Donor) |
| N41 | OE2 | GLU- 592 | 2.65 | 172.51 | H-Bond (Ligand Donor) |
| DuAr | CZ | ARG- 596 | 3.66 | 17.97 | Pi/Cation |
| C08 | CD | ARG- 596 | 3.92 | 0 | Hydrophobic |
| C18 | CG1 | VAL- 677 | 4.05 | 0 | Hydrophobic |
| C08 | CZ3 | TRP- 678 | 3.41 | 0 | Hydrophobic |
| C09 | CH2 | TRP- 678 | 3.62 | 0 | Hydrophobic |
| C18 | CE2 | TRP- 678 | 4.06 | 0 | Hydrophobic |
| C18 | CZ | PHE- 691 | 3.67 | 0 | Hydrophobic |