



2.600 Å
X-ray
2013-02-05
| Name: | Kynurenine 3-monooxygenase |
|---|---|
| ID: | KMO_YEAST |
| AC: | P38169 |
| Organism: | Saccharomyces cerevisiae |
| Reign: | Eukaryota |
| TaxID: | 559292 |
| EC Number: | / |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 100 % |
| B-Factor: | 19.550 |
|---|---|
| Number of residues: | 61 |
| Including | |
| Standard Amino Acids: | 58 |
| Non Standard Amino Acids: | 0 |
| Water Molecules: | 3 |
| Cofactors: | |
| Metals: | |
| Ligandability | Volume (Å3) |
|---|---|
| 0.667 | 1778.625 |
| % Hydrophobic | % Polar |
|---|---|
| 42.50 | 57.50 |
| According to VolSite | |
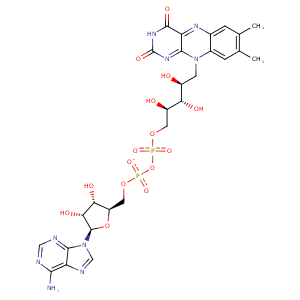
| HET Code: | FAD |
|---|---|
| Formula: | C27H31N9O15P2 |
| Molecular weight: | 783.534 g/mol |
| DrugBank ID: | DB03147 |
| Buried Surface Area: | 62.93 % |
| Polar Surface area: | 381.7 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 22 |
| H-Bond Donors: | 7 |
| Rings: | 6 |
| Aromatic rings: | 3 |
| Anionic atoms: | 2 |
| Cationic atoms: | 0 |
| Rule of Five Violation: | 3 |
| Rotatable Bonds: | 13 |
| X | Y | Z |
|---|---|---|
| 41.6756 | 7.55181 | 0.482245 |
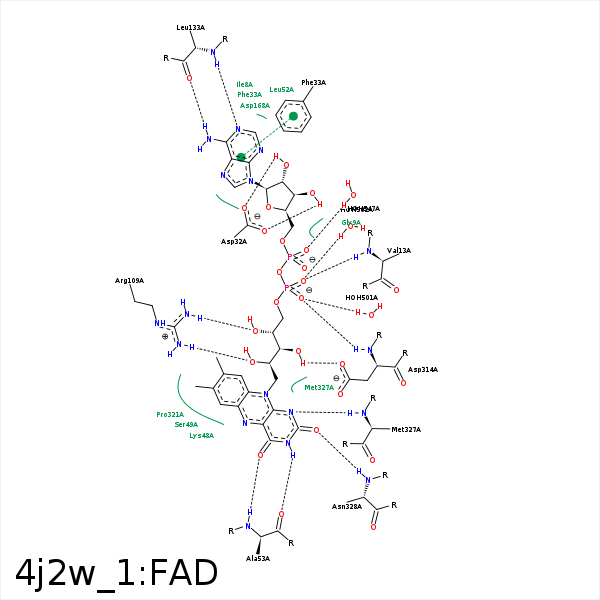
 Image generated by PoseView
Image generated by PoseView



Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| C5' | CG2 | VAL- 13 | 4.33 | 0 | Hydrophobic |
| O1P | N | VAL- 13 | 2.95 | 160.25 | H-Bond (Protein Donor) |
| O3B | OD2 | ASP- 32 | 2.81 | 177.76 | H-Bond (Ligand Donor) |
| O2B | OD1 | ASP- 32 | 2.84 | 172.29 | H-Bond (Ligand Donor) |
| N3A | N | PHE- 33 | 3.47 | 144.06 | H-Bond (Protein Donor) |
| C2B | CE2 | PHE- 33 | 4.01 | 0 | Hydrophobic |
| O3B | NE | ARG- 34 | 3.22 | 136.48 | H-Bond (Protein Donor) |
| C8M | CB | LYS- 48 | 3.73 | 0 | Hydrophobic |
| C7M | CB | SER- 49 | 3.87 | 0 | Hydrophobic |
| N3 | O | ALA- 53 | 3.2 | 157.77 | H-Bond (Ligand Donor) |
| O4 | N | ALA- 53 | 2.87 | 168.93 | H-Bond (Protein Donor) |
| O2' | NH1 | ARG- 109 | 3.35 | 140.45 | H-Bond (Protein Donor) |
| O2' | NH2 | ARG- 109 | 3.04 | 158.21 | H-Bond (Protein Donor) |
| O4' | NH1 | ARG- 109 | 3.27 | 147.96 | H-Bond (Protein Donor) |
| N6A | O | LEU- 133 | 3.29 | 150.98 | H-Bond (Ligand Donor) |
| N1A | N | LEU- 133 | 3.08 | 143.2 | H-Bond (Protein Donor) |
| C1B | CB | ASP- 168 | 4.4 | 0 | Hydrophobic |
| C7M | CE1 | TYR- 195 | 3.75 | 0 | Hydrophobic |
| C8M | CE2 | TYR- 195 | 3.85 | 0 | Hydrophobic |
| O3' | OD1 | ASP- 314 | 2.76 | 159.78 | H-Bond (Ligand Donor) |
| O3' | OD2 | ASP- 314 | 3.41 | 141.73 | H-Bond (Ligand Donor) |
| C5' | CB | ASP- 314 | 4.36 | 0 | Hydrophobic |
| O2P | N | ASP- 314 | 3.02 | 162.74 | H-Bond (Protein Donor) |
| C8 | CB | PRO- 321 | 3.9 | 0 | Hydrophobic |
| N1 | N | MET- 327 | 3.05 | 165.56 | H-Bond (Protein Donor) |
| C2' | CB | MET- 327 | 4.2 | 0 | Hydrophobic |
| C4' | CB | MET- 327 | 4.28 | 0 | Hydrophobic |
| O2 | N | ASN- 328 | 2.85 | 171.1 | H-Bond (Protein Donor) |
| O2P | O | HOH- 501 | 2.9 | 179.96 | H-Bond (Protein Donor) |
| O1P | O | HOH- 502 | 2.57 | 155.75 | H-Bond (Protein Donor) |
| O2A | O | HOH- 547 | 2.58 | 179.96 | H-Bond (Protein Donor) |