



2.300 Å
X-ray
2009-02-19
| Name: | Peroxisome proliferator-activated receptor gamma |
|---|---|
| ID: | PPARG_HUMAN |
| AC: | P37231 |
| Organism: | Homo sapiens |
| Reign: | Eukaryota |
| TaxID: | 9606 |
| EC Number: | / |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 100 % |
| B-Factor: | 44.500 |
|---|---|
| Number of residues: | 43 |
| Including | |
| Standard Amino Acids: | 43 |
| Non Standard Amino Acids: | 0 |
| Water Molecules: | 0 |
| Cofactors: | |
| Metals: | |
| Ligandability | Volume (Å3) |
|---|---|
| 1.593 | 877.500 |
| % Hydrophobic | % Polar |
|---|---|
| 65.77 | 34.23 |
| According to VolSite | |
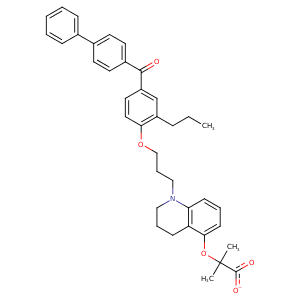
| HET Code: | 2PQ |
|---|---|
| Formula: | C38H40NO5 |
| Molecular weight: | 590.728 g/mol |
| DrugBank ID: | - |
| Buried Surface Area: | 69.13 % |
| Polar Surface area: | 78.9 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 6 |
| H-Bond Donors: | 0 |
| Rings: | 5 |
| Aromatic rings: | 4 |
| Anionic atoms: | 1 |
| Cationic atoms: | 0 |
| Rule of Five Violation: | 2 |
| Rotatable Bonds: | 13 |
| X | Y | Z |
|---|---|---|
| 36.5965 | -5.81245 | 38.9653 |
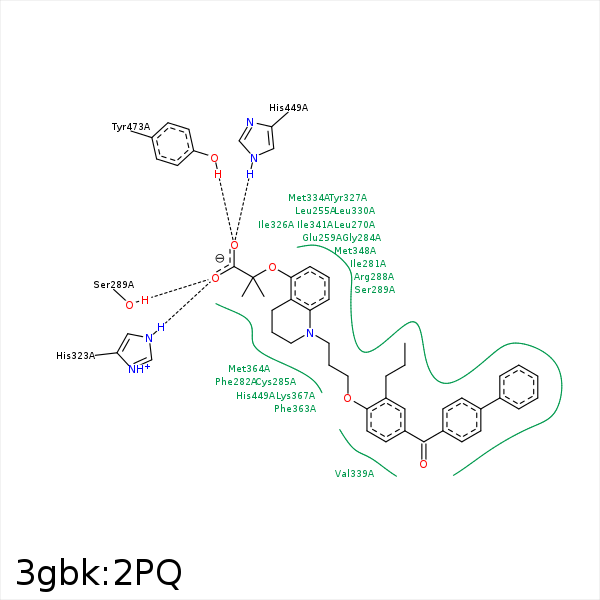
 Image generated by PoseView
Image generated by PoseView



Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| CAD | CD1 | LEU- 255 | 3.64 | 0 | Hydrophobic |
| CAD | CG | GLU- 259 | 3.29 | 0 | Hydrophobic |
| CAI | CD2 | LEU- 270 | 4.36 | 0 | Hydrophobic |
| CAZ | CD1 | PHE- 282 | 4.16 | 0 | Hydrophobic |
| CBA | CE1 | PHE- 282 | 3.63 | 0 | Hydrophobic |
| CBF | CZ | PHE- 282 | 3.35 | 0 | Hydrophobic |
| CAL | SG | CYS- 285 | 4.25 | 0 | Hydrophobic |
| CAZ | SG | CYS- 285 | 3.87 | 0 | Hydrophobic |
| CBG | CB | CYS- 285 | 4.46 | 0 | Hydrophobic |
| CBA | CB | CYS- 285 | 4.16 | 0 | Hydrophobic |
| CAM | SG | CYS- 285 | 3.92 | 0 | Hydrophobic |
| CBF | CG | GLN- 286 | 4.05 | 0 | Hydrophobic |
| CBG | CG | GLN- 286 | 3.62 | 0 | Hydrophobic |
| CAO | CG | ARG- 288 | 3.89 | 0 | Hydrophobic |
| CBN | CG | ARG- 288 | 3.78 | 0 | Hydrophobic |
| CBG | CB | SER- 289 | 4.06 | 0 | Hydrophobic |
| CBO | CB | SER- 289 | 3.96 | 0 | Hydrophobic |
| CAS | CB | SER- 289 | 4.09 | 0 | Hydrophobic |
| OBC | OG | SER- 289 | 2.97 | 162.17 | H-Bond (Protein Donor) |
| OBC | NE2 | HIS- 323 | 2.97 | 147.45 | H-Bond (Protein Donor) |
| CBO | CG2 | ILE- 326 | 3.32 | 0 | Hydrophobic |
| CAT | CG2 | ILE- 326 | 4.14 | 0 | Hydrophobic |
| CBH | CD2 | LEU- 330 | 4.25 | 0 | Hydrophobic |
| CBI | CD2 | LEU- 330 | 3.41 | 0 | Hydrophobic |
| CBM | CD1 | LEU- 330 | 3.77 | 0 | Hydrophobic |
| CBK | CE | MET- 334 | 4.36 | 0 | Hydrophobic |
| CAR | CG1 | VAL- 339 | 4.12 | 0 | Hydrophobic |
| CBK | CG1 | VAL- 339 | 4.43 | 0 | Hydrophobic |
| CAI | CD1 | ILE- 341 | 4.16 | 0 | Hydrophobic |
| CAH | CB | ILE- 341 | 4.16 | 0 | Hydrophobic |
| CAG | CG2 | ILE- 341 | 4.17 | 0 | Hydrophobic |
| CAQ | CG2 | ILE- 341 | 3.25 | 0 | Hydrophobic |
| CAK | CE | MET- 348 | 4.31 | 0 | Hydrophobic |
| CBK | CD1 | LEU- 353 | 4.43 | 0 | Hydrophobic |
| CBA | CD1 | PHE- 363 | 3.89 | 0 | Hydrophobic |
| CAY | CB | PHE- 363 | 3.7 | 0 | Hydrophobic |
| CAY | SD | MET- 364 | 3.69 | 0 | Hydrophobic |
| CAZ | CE | MET- 364 | 3.9 | 0 | Hydrophobic |
| CBJ | SD | MET- 364 | 3.67 | 0 | Hydrophobic |
| CBJ | CB | LYS- 367 | 4.43 | 0 | Hydrophobic |
| CBI | CG | LYS- 367 | 4.16 | 0 | Hydrophobic |
| OBE | NE2 | HIS- 449 | 3.34 | 126.97 | H-Bond (Protein Donor) |
| OBR | NE2 | HIS- 449 | 2.88 | 143.81 | H-Bond (Protein Donor) |
| CBF | CD1 | LEU- 453 | 4.32 | 0 | Hydrophobic |
| CBF | CD2 | LEU- 465 | 3.93 | 0 | Hydrophobic |
| CBG | CD1 | LEU- 469 | 4.01 | 0 | Hydrophobic |
| OBR | OH | TYR- 473 | 2.7 | 127.61 | H-Bond (Protein Donor) |