



1.980 Å
X-ray
2007-12-13
| Name: | Pteridine reductase |
|---|---|
| ID: | O76290_TRYBB |
| AC: | O76290 |
| Organism: | Trypanosoma brucei brucei |
| Reign: | Eukaryota |
| TaxID: | 5702 |
| EC Number: | / |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 100 % |
| B-Factor: | 23.654 |
|---|---|
| Number of residues: | 17 |
| Including | |
| Standard Amino Acids: | 16 |
| Non Standard Amino Acids: | 1 |
| Water Molecules: | 0 |
| Cofactors: | NAP |
| Metals: | |
| Ligandability | Volume (Å3) |
|---|---|
| 0.749 | 914.625 |
| % Hydrophobic | % Polar |
|---|---|
| 44.65 | 55.35 |
| According to VolSite | |
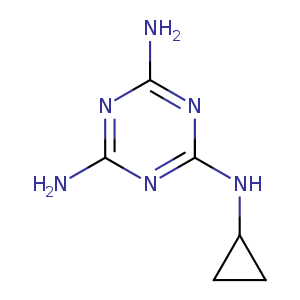
| HET Code: | AX3 |
|---|---|
| Formula: | C6H10N6 |
| Molecular weight: | 166.184 g/mol |
| DrugBank ID: | - |
| Buried Surface Area: | 69.07 % |
| Polar Surface area: | 102.74 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 6 |
| H-Bond Donors: | 3 |
| Rings: | 2 |
| Aromatic rings: | 1 |
| Anionic atoms: | 0 |
| Cationic atoms: | 0 |
| Rule of Five Violation: | 0 |
| Rotatable Bonds: | 2 |
| X | Y | Z |
|---|---|---|
| 9.98175 | -10.5649 | 14.5425 |
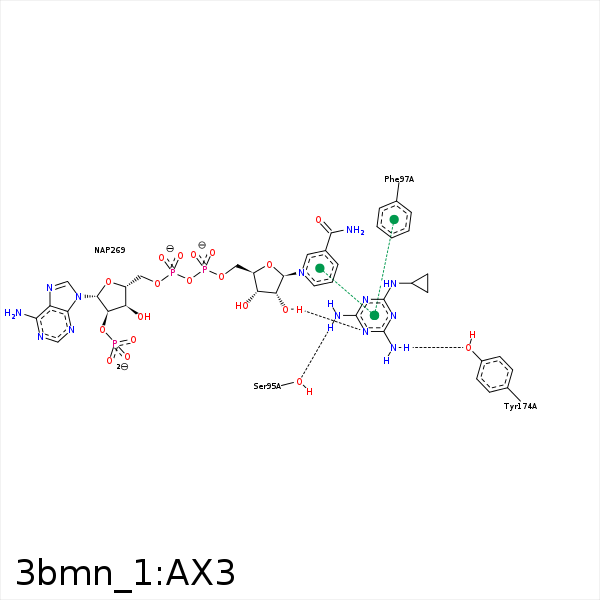
 Image generated by PoseView
Image generated by PoseView



Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| NAB | O | SER- 95 | 3.11 | 121.56 | H-Bond (Ligand Donor) |
| NAB | OG | SER- 95 | 2.6 | 161.45 | H-Bond (Ligand Donor) |
| CAD | CD1 | PHE- 97 | 3.8 | 0 | Hydrophobic |
| NAA | OH | TYR- 174 | 2.52 | 164.95 | H-Bond (Ligand Donor) |
| CAC | CB | PRO- 210 | 3.45 | 0 | Hydrophobic |
| NAE | O2D | NAP- 269 | 2.88 | 149.98 | H-Bond (Protein Donor) |
| NAB | O1A | NAP- 269 | 3.29 | 133.14 | H-Bond (Ligand Donor) |