



2.300 Å
X-ray
2006-05-24
| Min | Mean | Median | Standard Deviation | Max | Count | |
|---|---|---|---|---|---|---|
| pChEMBL: | 5.500 | 5.630 | 5.500 | 0.180 | 5.890 | 3 |
| Name: | Tyrosine-protein phosphatase non-receptor type 1 |
|---|---|
| ID: | PTN1_HUMAN |
| AC: | P18031 |
| Organism: | Homo sapiens |
| Reign: | Eukaryota |
| TaxID: | 9606 |
| EC Number: | 3.1.3.48 |
| Chain Name: | Percentage of Residues within binding site |
|---|---|
| A | 100 % |
| B-Factor: | 59.810 |
|---|---|
| Number of residues: | 26 |
| Including | |
| Standard Amino Acids: | 25 |
| Non Standard Amino Acids: | 0 |
| Water Molecules: | 1 |
| Cofactors: | |
| Metals: | |
| Ligandability | Volume (Å3) |
|---|---|
| 0.308 | 239.625 |
| % Hydrophobic | % Polar |
|---|---|
| 59.15 | 40.85 |
| According to VolSite | |
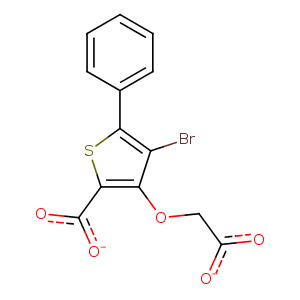
| HET Code: | 509 |
|---|---|
| Formula: | C13H7BrO5S |
| Molecular weight: | 355.161 g/mol |
| DrugBank ID: | DB07130 |
| Buried Surface Area: | 66.5 % |
| Polar Surface area: | 117.73 Å2 |
| Number of | |
|---|---|
| H-Bond Acceptors: | 5 |
| H-Bond Donors: | 0 |
| Rings: | 2 |
| Aromatic rings: | 2 |
| Anionic atoms: | 2 |
| Cationic atoms: | 0 |
| Rule of Five Violation: | 0 |
| Rotatable Bonds: | 5 |
| X | Y | Z |
|---|---|---|
| 45.8668 | 13.816 | 1.87765 |
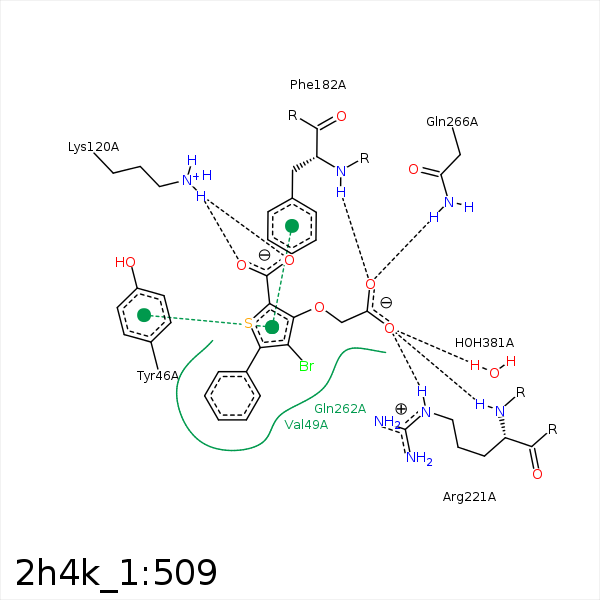
 Image generated by PoseView
Image generated by PoseView



Represent the protein/ligand binding mode, centered on the ligand
Dashed lines represents hydrogen bonds and metal interactions
Green residue labels for amino acids with hydrophobic contacts (green lines) to the ligand

| Ligand | Protein | Interaction | |||
|---|---|---|---|---|---|
| Atom | Atom | Residue | Distance (Å) | Angle (°) | Type |
| C3 | CB | TYR- 46 | 4.15 | 0 | Hydrophobic |
| C1 | CB | ASP- 48 | 4.17 | 0 | Hydrophobic |
| C2 | CG2 | VAL- 49 | 3.54 | 0 | Hydrophobic |
| O1 | NZ | LYS- 120 | 2.71 | 130.1 | H-Bond (Protein Donor) |
| O2 | NZ | LYS- 120 | 3.09 | 158.18 | H-Bond (Protein Donor) |
| O1 | NZ | LYS- 120 | 2.71 | 0 | Ionic (Protein Cationic) |
| O2 | NZ | LYS- 120 | 3.09 | 0 | Ionic (Protein Cationic) |
| C12 | CE2 | PHE- 182 | 3.75 | 0 | Hydrophobic |
| BR1 | CZ | PHE- 182 | 4.35 | 0 | Hydrophobic |
| O4 | N | PHE- 182 | 2.8 | 148.87 | H-Bond (Protein Donor) |
| BR1 | CB | ALA- 217 | 4 | 0 | Hydrophobic |
| BR1 | CG1 | ILE- 219 | 3.52 | 0 | Hydrophobic |
| O5 | N | ARG- 221 | 3.03 | 159.36 | H-Bond (Protein Donor) |
| O5 | NE | ARG- 221 | 3.33 | 155.41 | H-Bond (Protein Donor) |
| BR1 | CB | GLN- 262 | 4.27 | 0 | Hydrophobic |
| O4 | NE2 | GLN- 266 | 3 | 151.87 | H-Bond (Protein Donor) |
| O5 | O | HOH- 381 | 2.76 | 179.97 | H-Bond (Protein Donor) |